Computational Methods in Drug Discovery
Published online:
2014 Jan. doi: 10.1124/pr.112.007336
PMID:
What is this article about:
Computational Methods in Drug Discovery, Computer Aided Drug Discovery, structure based or ligand-based methods, molecular descriptors.
Editors:
Gregory Sliwoski, Sandeepkumar Kothiwale, Jens Meiler, and Edward W. Lowe, Jr.
Associate Editor:
Eric L. Barker
Structure-Based Virtual high-throughput screening, Quantitative structure activity relationship
Abstract
Computer-aided drug discovery/design methods have played a major role in the development of therapeutically important small molecules for over three decades. These methods are broadly classified as either structure-based or ligand-based methods. Structure-based methods are in principle analogous to high-throughput screening in that both target and ligand structure information is imperative. Structure-based approaches include ligand docking, pharmacophore, and ligand design methods. The article discusses theory behind the most important methods and recent successful applications. Ligand-based methods use only ligand information for predicting activity depending on its similarity/dissimilarity to previously known active ligands. We review widely used ligand-based methods such as ligand-based pharmacophores, molecular descriptors, and quantitative structure-activity relationships. In addition, important tools such as target/ligand data bases, homology modeling, ligand fingerprint methods, etc., necessary for successful implementation of various computer-aided drug discovery/design methods in a drug discovery campaign are discussed. Finally, computational methods for toxicity prediction and optimization for favorable physiologic properties are discussed with successful examples from literature.
Introduction
On October 5, 1981, Fortune magazine published a cover article entitled the “Next Industrial Revolution: Designing Drugs by Computer at Merck” (Van Drie, 2007). Some have credited this as being the start of intense interest in the potential for computer-aided drug design (CADD). Although progress was being made in CADD, the potential for high-throughput screening (HTS) had begun to take precedence as a means for finding novel therapeutics. This brute force approach relies on automation to screen high numbers of molecules in search of those that elicit the desired biologic response. The method has the advantage of requiring minimal compound design or prior knowledge, and technologies required to screen large libraries have become more efficient. However, although traditional HTS often results in multiple hit compounds, some of which are capable of being modified into a lead and later a novel therapeutic, the hit rate for HTS is often extremely low. This low hit rate has limited the usage of HTS to research programs capable of screening large compound libraries. In the past decade, CADD has reemerged as a way to significantly decrease the number of compounds necessary to screen while retaining the same level of lead compound discovery. Many compounds predicted to be inactive can be skipped, and those predicted to be active can be prioritized. This reduces the cost and workload of a full HTS screen without compromising lead discovery. Additionally, traditional HTS assays often require extensive development and validation before they can be used. Because CADD requires significantly less preparation time, experimenters can perform CADD studies while the traditional HTS assay is being prepared. The fact that both of these tools can be used in parallel provides an additional benefit for CADD in a drug discovery project.
For example, researchers at Pharmacia (now part of Pfizer) used CADD tools to screen for inhibitors of tyrosine phosphatase-1B, an enzyme implicated in diabetes. Their virtual screen yielded 365 compounds, 127 of which showed effective inhibition, a hit rate of nearly 35%. Simultaneously, this group performed a traditional HTS against the same target. Of the 400,000 compounds tested, 81 showed inhibition, producing a hit rate of only 0.021%. This comparative case effectively displays the power of CADD (Doman et al., 2002). CADD has already been used in the discovery of compounds that have passed clinical trials and become novel therapeutics in the treatment of a variety of diseases. Some of the earliest examples of approved drugs that owe their discovery in large part to the tools of CADD include the following: carbonic anhydrase inhibitor dorzolamide, approved in 1995 (Vijayakrishnan 2009); the angiotensin-converting enzyme (ACE) inhibitor captopril, approved in 1981 as an antihypertensive drug (Talele et al., 2010); three therapeutics for the treatment of human immunodeficiency virus (HIV): saquinavir (approved in 1995), ritonavir, and indinavir (both approved in 1996) (Van Drie 2007); and tirofiban, a fibrinogen antagonist approved in 1998 (Hartman et al., 1992).
One of the most striking examples of the possibilities presented from CADD occurred in 2003 with the search for novel transforming growth factor-β1 receptor kinase inhibitors. One group at Eli Lilly used a traditional HTS to identify a lead compound that was subsequently improved by examination of structure-activity relationship using in vitro assays (Sawyer et al., 2003), whereas a group at Biogen Idec used a CADD approach involving virtual HTS based on the structural interactions between a weak inhibitor and transforming growth factor-β1 receptor kinase (Singh et al., 2003a). Upon the virtual screening of compounds, the group at Biogen Idec identified 87 hits, the best hit being identical in structure to the lead compound discovered through the traditional HTS approach at Eli Lilly (Shekhar 2008). In this situation, CADD, a method involving reduced cost and workload, was capable of producing the same lead as a full-scale HTS (Fig. 1) (Sawyer et al., 2003).
Introduction
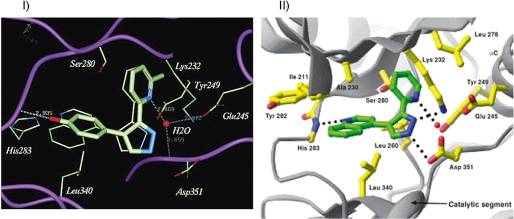
Identical lead compounds are discovered in a traditional high-throughput screen and structure-based virtual high-throughput screen. I, X-ray crystal structures of 1 and 18 bound to the ATP-binding site of the TβR-I kinase domain discovered using traditional high-throughput screening. Compound 1, shown as the thinner wire-frame is the original hit from the HTS and is identical to that which was discovered using virtual screening. Compound 18 is a higher affinity compound after lead optimization. II, X-ray crystal structure of compound HTS466284 bound to the TβRI active site. This compound is identical to compound 1 in I but was discovered using structure-based virtual high-throughput screening.
A. Position of Computer-Aided Drug Design in the Drug Discovery Pipeline
CADD is capable of increasing the hit rate of novel drug compounds because it uses a much more targeted search than traditional HTS and combinatorial chemistry. It not only aims to explain the molecular basis of therapeutic activity but also to predict possible derivatives that would improve activity. In a drug discovery campaign, CADD is usually used for three major purposes: (1) filter large compound libraries into smaller sets of predicted active compounds that can be tested experimentally; (2) guide the optimization of lead compounds, whether to increase its affinity or optimize drug metabolism and pharmacokinetics (DMPK) properties including absorption, distribution, metabolism, excretion, and the potential for toxicity (ADMET); (3) design novel compounds, either by “growing” starting molecules one functional group at a time or by piecing together fragments into novel chemotypes. Figure 2 illustrates the position of CADD in drug discovery pipeline.
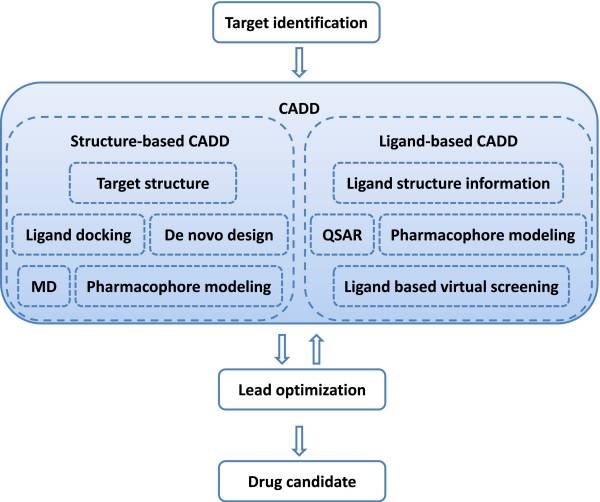
CADD in drug discovery/design pipeline. A therapeutic target is identified against which a drug has to be developed. Depending on the availability of structure information, a structure-based approach or a ligand-based approach is used. A successful CADD campaign will allow identification of multiple lead compounds. Lead identification is often followed by several cycles of lead optimization and subsequent lead identification using CADD. Lead compounds are tested in vivo to identify drug candidates.
CADD can be classified into two general categories: structure-based and ligand-based. Structure-based CADD relies on the knowledge of the target protein structure to calculate interaction energies for all compounds tested, whereas ligand-based CADD exploits the knowledge of known active and inactive molecules through chemical similarity searches or construction of predictive, quantitative structure-activity relation (QSAR) models (Kalyaanamoorthy and Chen, 2011). Structure-based CADD is generally preferred where high-resolution structural data of the target protein are available, i.e., for soluble proteins that can readily be crystallized. Ligand-based CADD is generally preferred when no or little structural information is available, often for membrane protein targets. The central goal of structure-based CADD is to design compounds that bind tightly to the target, i.e., with large reduction in free energy, improved DMPK/ADMET properties, and are target specific, i.e., have reduced off-target effects (Jorgensen, 2010). A successful application of these methods will result in a compound that has been validated in vitro and in vivo and its binding location has been confirmed, ideally through a cocrystal structure.
One of the most common uses in CADD is the screening of virtual compound libraries, also known as virtual high-throughput screening (vHTS). This allows experimentalists to focus resources on testing compounds likely to have any activity of interest. In this way, a researcher can identify an equal number of hits while screening significantly less compounds, because compounds predicted to be inactive with high confidence may be skipped. Avoiding a large population of inactive compounds saves money and time, because the size of the experimental HTS is significantly reduced without sacrificing a large degree of hits. Ripphausen et al. (2010) note that the first mention of vHTS was in 1997 (Horvath, 1997) and chart an increasing rate of publication for the application of vHTS between 1997 and 2010. They also found that the largest fraction of hits has been obtained for G-protein-coupled receptors (GPCRs) followed by kinases (Ripphausen et al., 2010).
vHTS comes in many forms, including chemical similarity searches by fingerprints or topology, selecting compounds by predicted biologic activity through QSAR models or pharmacophore mapping, and virtual docking of compounds into target of interest, known as structure-based docking (Enyedy and Egan, 2008). These methods allow the ranking of “hits” from the virtual compound library for acquisition. The ranking can reflect a property of interest such as percent similarity to a query compound or predicted biologic activity, or in the case of docking, the lowest energy scoring poses for each ligand bound to the target of interest (Joffe, 1991). Often initial hits are rescored and ranked using higher level computational techniques that are too time consuming to be applied to full-scale vHTS. It is important to note that vHTS does not aim to identify a drug compound that is ready for clinical testing, but rather to find leads with chemotypes that have not previously been associated with a target. This is not unlike a traditional HTS where a compound is generally considered a hit if its activity is close to 10 µM. Through iterative rounds of chemical synthesis and in vitro testing, a compound is first developed into a “lead” with higher affinity, some understanding of its structure-activity-relation, and initial tests for DMPK/ADMET properties. Only after further iterative rounds of lead-to-drug optimization and in vivo testing does a compound reach a clinically appropriate potency and acceptable DMPK/ADMET properties (Jorgensen, 2004). For example, the literature survey performed by Ripphausen et al. (2010) revealed that a majority of successful vHTS applications identified a small number of hits that are usually active in the micromolar range, and hits with low nanomolar potency are only rarely identified.
The cost benefit of using computational tools in the lead optimization phase of drug development is substantial. Development of new drugs can cost anywhere in the range of 400 million to 2 billion dollars, with synthesis and testing of lead analogs being a large contributor to that sum (Basak, 2012). Therefore, it is beneficial to apply computational tools in hit-to-lead optimization to cover a wider chemical space while reducing the number of compounds that must be synthesized and tested in vitro. The computational optimization of a hit compound can involve a structure-based analysis of docking poses and energy profiles for hit analogs, ligand-based screening for compounds with similar chemical structure or improved predicted biologic activity, or prediction of favorable DMPK/ADMET properties. The comparably low cost of CADD compared with chemical synthesis and biologic characterization of compounds make these methods attractive to focus, reduce, and diversify the chemical space that is explored (Enyedy and Egan, 2008).
De novo drug design is another tool in CADD methods, but rather than screening libraries of previously synthesized compounds, it involves the design of novel compounds. A structure generator is needed to sample the space of chemicals. Given the size of the search space (more than 1060 molecules) (Bohacek et al., 1996) heuristics are used to focus these algorithms on molecules that are predicted to be highly active, readily synthesizable, devoid of undesirable properties, often derived from a starting scaffold with demonstrated activity, etc. Additionally, effective sampling strategies are used while dealing with large search spaces such as evolutionary algorithms, metropolis search, or simulated annealing (Schneider et al., 2009). The construction algorithms are generally defined as either linking or growing techniques. Linking algorithms involve docking of small fragments or functional groups such as rings, acetyl groups, esters, etc., to particular binding sites followed by linking fragments from adjacent sites. Growing algorithms, on the other hand, begin from a single fragment placed in the binding site to which fragments are added, removed, and changed to improve activity. Similar to vHTS, the role of de novo drug design is not to design the single compound with nanomolar activity and acceptable DMPK/ADMET properties but rather to design a lead compound that can be subsequently improved.
B. Ligand Databases for Computer-Aided Drug Design
Virtual HTS uses high-performance computing to screen large chemical data bases and prioritize compounds for synthesis. Current hardware and algorithms allow structure-based screening of up to 100,000 molecules per day using parallel processing clusters (Agarwal and Fishwick, 2010). To perform a virtual screen, however, a virtual library must be available for screening. Virtual libraries can be acquired in a variety of sizes and designs including general libraries that can be used to screen against any target, focused libraries that are designed for a family of related targets, and targeted libraries that are specifically designed for a single target (Takahashi et al., 2011).
General libraries can be constructed using a variety of computational and combinatorial tools. Early systems used molecular formula as the only constraint for structure generation, resulting in all possible structures for a predetermined limit in the number of atoms. As comprehensive computational enumeration of all chemical space is and will remain infeasible, additional restrictions are applied. Typically, chemical entities difficult to synthesize or known/expected to cause unfavorable DMPK/ADMET properties are excluded. Fink et al. proposed a generation method for the construction of virtual libraries that involved the use of connected graphs populated with C, N, O, and F atoms and pruned based on molecular structure constraints and the removal of unstable structures. The final data base proposed with this method is called the GDB (Generated a DataBase) and contains 26.4 million chemical structures that have been used for vHTS (Fink et al., 2005; Fink and Reymond, 2007). A more recent variation of this data base called GDB-13 includes atoms C, N, O, S, and Cl (F is not included in this variation to accelerate computation) and contains 970 million compounds (Blum and Reymond, 2009).
Most frequently, vHTS focuses on drug-like molecules that have been synthesized or can be easily derived from already available starting material. For this purpose several small molecule data bases are available that provide a variety of information including known/available chemical compounds, drugs, carbohydrates, enzymes, reactants, and natural products (Ortholand and Ganesan, 2004; Song et al., 2009). Some widely used data bases are listed in Table 1.
This is only a very brief portion of the complete article. Please continue reading using the link below.
Wrote or contributed to the writing of the manuscript: Sliwoski, Kothiwale, Meiler, Lowe.
Start using Alphaperrin Computational™ Lab Environment
Read The Full Publication on PubMed.com

Discovering. Improving.
Drug Design Perfected.
Quick Links
A Guide To in silico Drug Design
Computational Methods in Drug Discovery
P: 910.756.6106
F: 279.300.3241